Bacterial genes are often found in an operon.

An operon is essentially an assembly line of regulatory DNA genes, controlled by a region called the ‘promoter’. The regulatory DNA sequences act as binding sites for regulatory proteins, that promote or inhibit transcription.
Operons are quite efficient, however, if a singular mutation occurs at any point on the operon, the entire polycistronic pathway can be impacted. Operons can also struggle to take advantage of environmental changes; all genes are activated when the promoter is active.
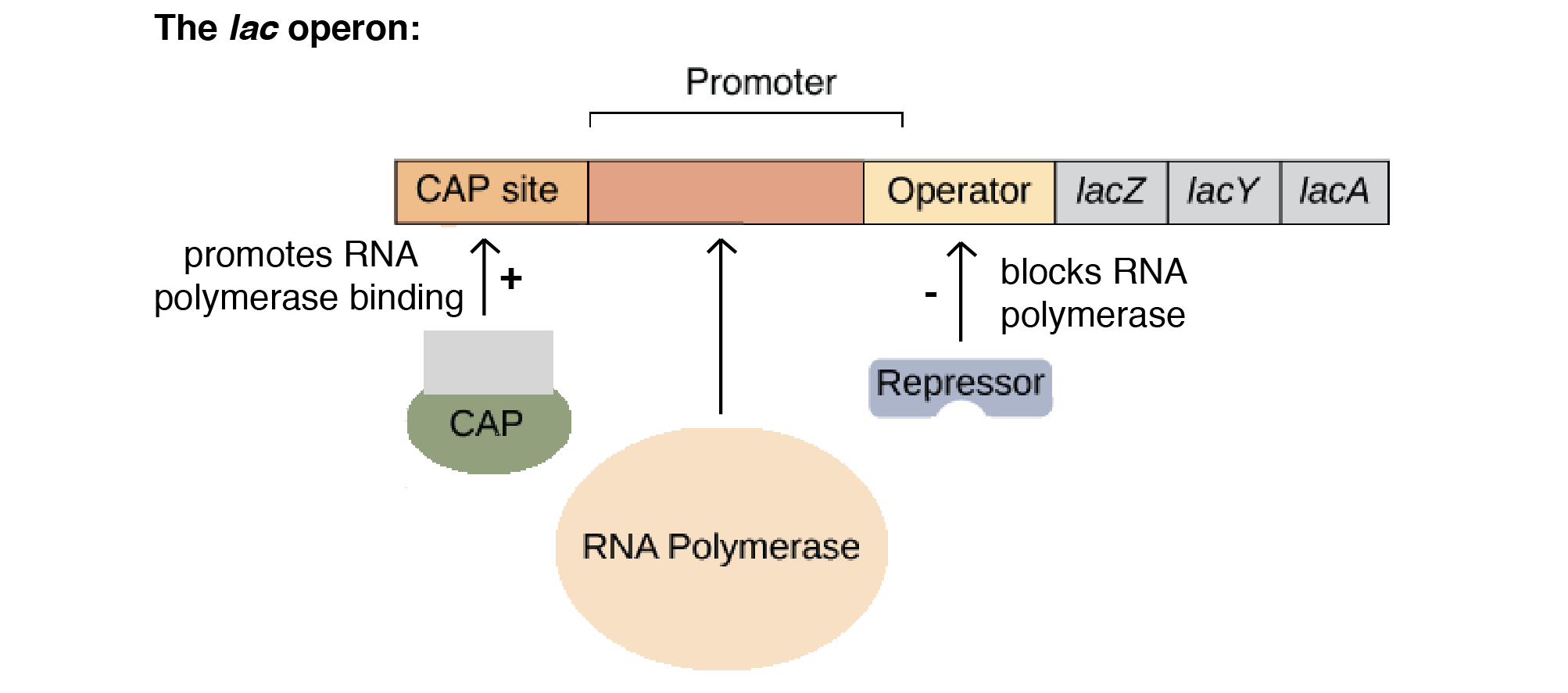
A common example of an operon is the lac operon.
The lactose, or lac, operon is most commonly found in the bacterium Escherichia coli (E. coli). The lac operon refers to a cluster of three structural genes that each encode for proteins involved in lactose metabolism. These genes are ‘lacA’, ‘lacY’ and ‘lacZ’. LacZ and lacY are essential for the utilisation of lactose by E. coli.
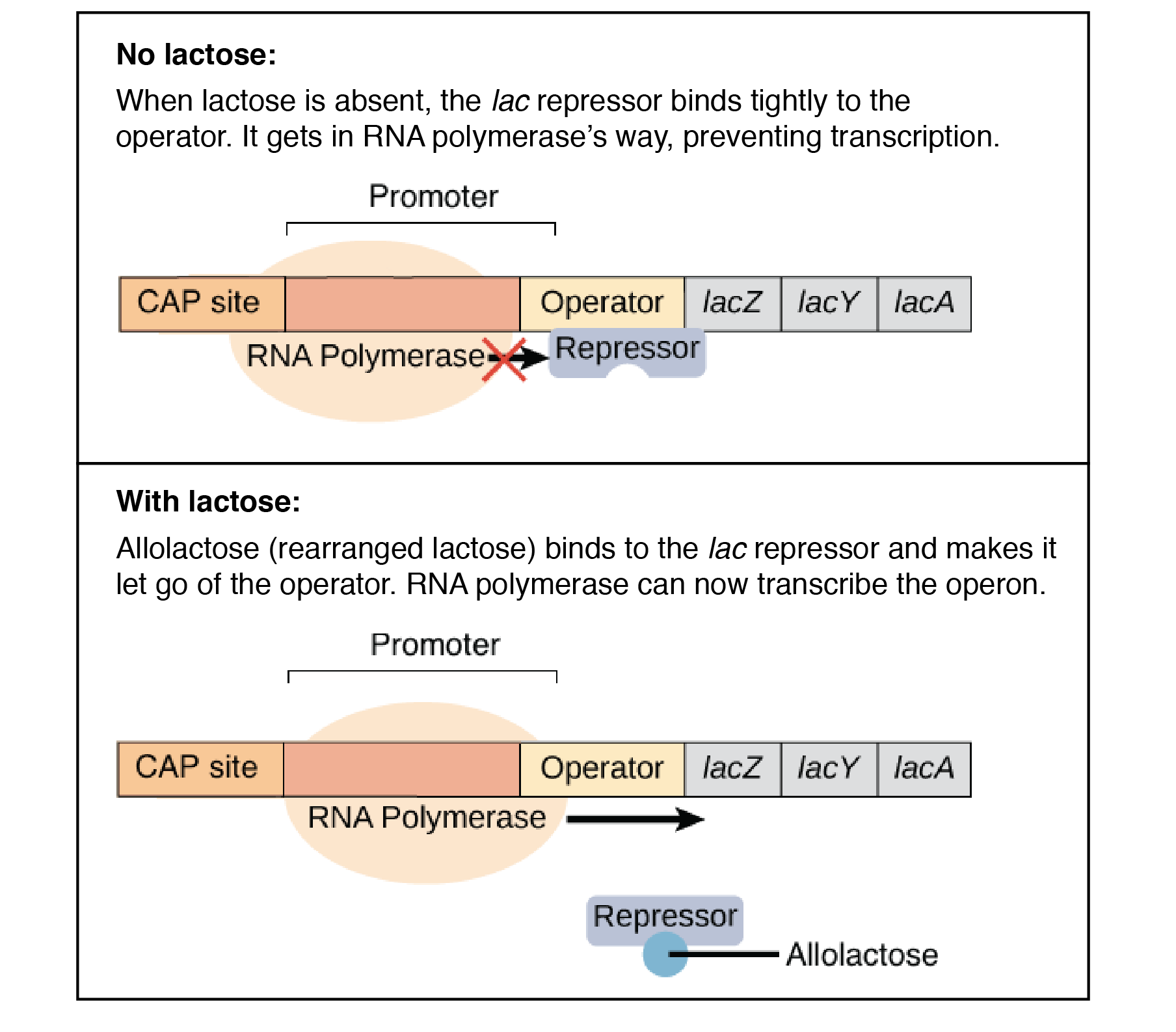
The lac operon is a negative, inducible system. When no lactose is present, a repressor binds to the operator – preventing transcription. In the presence of lactose, allolactose binds and inactivates the lac repressor. This allows RNA polymerase to bind to the lac operon – enabling transcription.
It is important to note that a repressor binds to the operator, which prevents RNA polymerase from binding and transcription occurring. This is negative control.
An activator encourages polymerase to bind to the promoter. This is positive control.
The lacA gene encodes for lactose transacetylase; an enzyme that transfers an acetyl group from acetyl-CoA to galactosides.
The lacY gene encodes for lactose permease; a transmembrane protein that facilitates the movement of lactose across the phospholipid bilayers that surround all cells and organelles via active transport. When glucose is present, lactose permease is not produced – hence, lactose cannot be transported into the cell.
The lacZ gene encodes for β-galactosidase; a bacterial enzyme that catalyses the breakdown of lactose into its component simple monosaccharides, glucose and galactose. The synthesis of β-galactosidase is activated when glucose levels are low, and lactose is present. When glucose is low, β-galactosidase and lactose fit together. Once together, a change in conformation of the enzyme occurs. This new conformation causes bond strain between the monosaccharides, until eventually the bond breaks, and glucose and galactose dissociate from the enzyme to provide energy to the bacterial system. β-galactosidase synthesis stops when glucose levels are sufficient.
Lactose permease actively transports lactose into the cell. Following this, β-galactosidase breaks down the lactose into its components galactose and glucose. β-galactosidase also converts lactose into allolactose, then converts the allolactose into galactose and glucose.
Catabolite repression regulates the lac operon via positive control. It is the process of glucose repression. There is an inverse relationship between glucose and cyclic-AMP (cAMP); when cellular glucose levels are high, cAMP is low, and vice versa. When cAMP is present, a catabolite activator protein (CAP) binds to the lac operon promoter, facilitating the binding or RNA polymerase to the promoter, leading to enhanced transcription of the operon’s genes.
Another common operon example is the tryptophan, or trp, operon. The trp operon is an example for negative repressible transcription.
When tryptophan is low, the inactive regulator protein (repressor) does not bind to the operator, enabling transcription. However, when tryptophan is high, the repressor and tryptophan bind together, then bind to the operator. This prevents transcription from occurring.
Attenuation is a mechanism for reducing expression of the trp operon when levels of tryptophan are high. Rather than blocking initiation of transcription, attenuation prevents completion of transcription. The attenuation of the trp operon works through a mechanism that depends on coupling (the translation of an mRNA that is still in the process of being transcribed).
The trp RNA is able to form a hairpin. When sections 1 and 2 pair, and 3 and 4 align, transcription is terminated. However, when 2 and 3 bind, transcription still occurs. This determines which regions pair up.
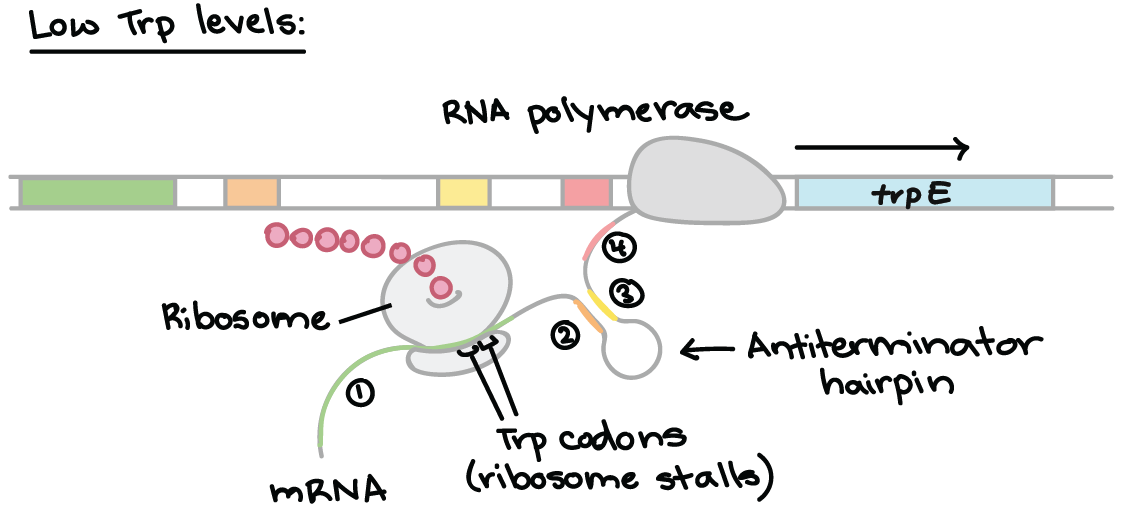
When tryptophan levels are low, the ribosome stalls at the trp codons in region 1. Region 2 then is not covered by the ribosome, where region 3 is transcribed. When region 3 is transcribed, it pairs with region 2 – the attenuator never forms and transcription continues.
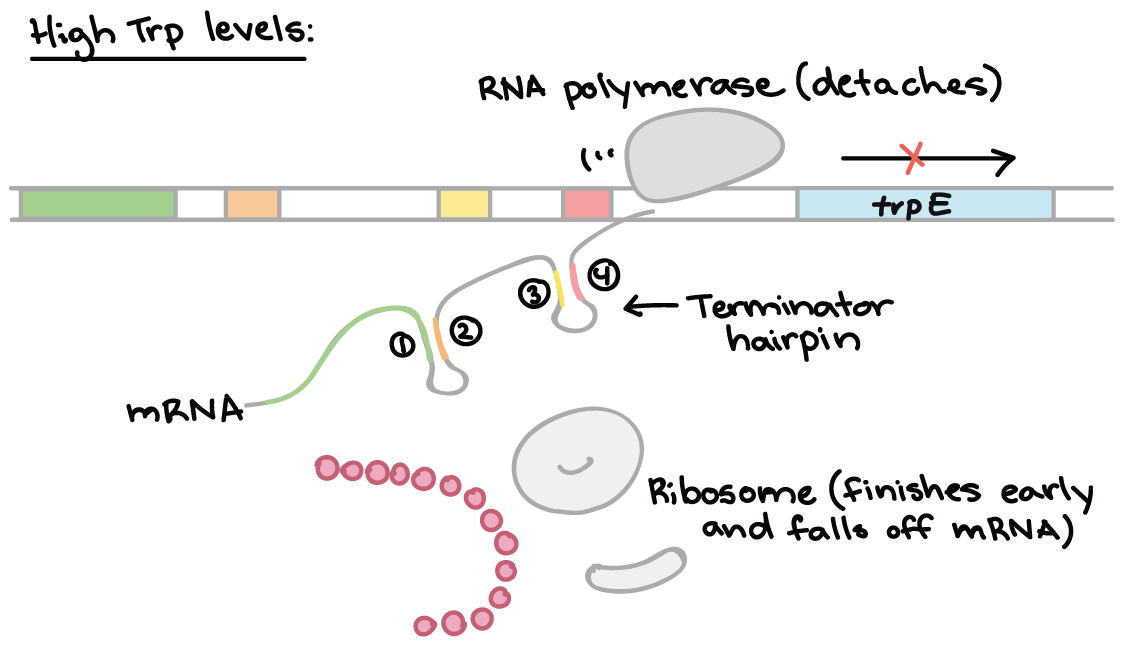
When tryptophan levels are high, RNA polymerase begins transcribing DNA – producing region 1 of the 5′ UTR. A ribosome binds to the 5′ end of the 5′ UTR, and translates region 1 while region 2 is being transcribed.
RNA polymerase transcribes region 3. The ribosome does not stall at the trp codons, because tryptophan is abundant.
The ribosome covers part of region 2, preventing pairing with region 3. Region 4 is transcribed and pairs with region 3, producing the attenuator that terminates transcription.